Choosing the correct BLAST algorithm
SequenceServer has an auto-detection feature that selects the appropriate BLAST algorithm for your input data and databases.
However, there are five basic BLAST algorithms: blastp, blastn, tblastx, tblastn, and blastx. Each algorithm has a different use case, and it’s essential to choose the appropriate one for your analysis. This post will help you choose the right one.
The appropriate BLAST algorithm choice depends on what you’re trying to do.
As biologists, we work with nucleotide sequences and protein (i.e., amino-acid) sequences. Several versions of BLAST exist so we can analyze both types of sequences. Are we searching with a nucleotide sequence or a protein sequence? Are we comparing that to a database of amino-acid sequences such as UniRef90 or to a database of nucleotide sequences such as the Telomere-to-Telomere human genome?
The correct BLAST algorithm depends on the type of query sequence and the type of database sequence. Below is a summary overview from our 2019 Mol Biol Evol paper:
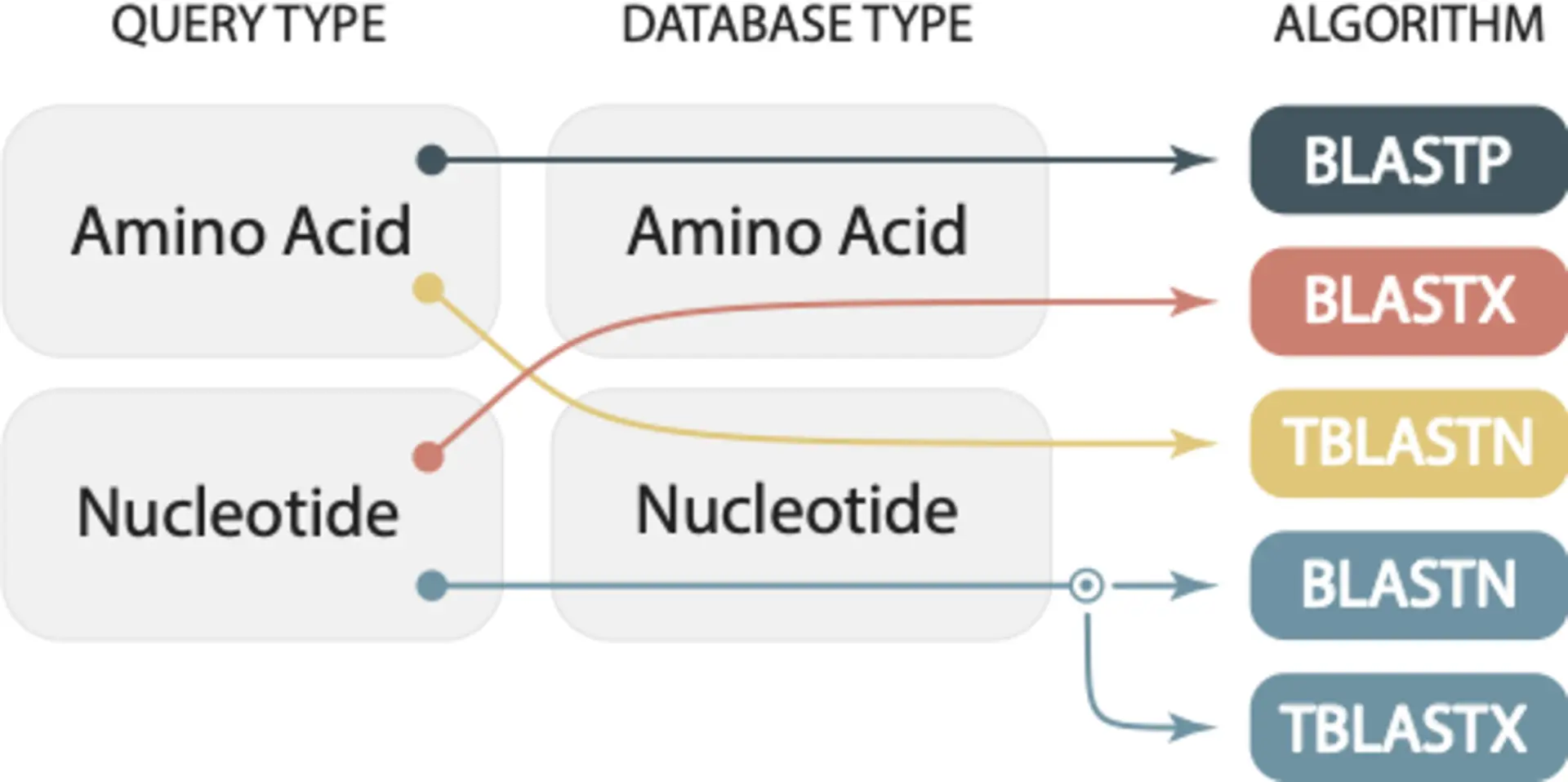
Choosing the wrong algorithm can lead to incorrect results
Choosing the wrong algorithm can lead to incorrect results. For example, if you want to search with a nucleotide query sequence but run blastp, BLAST will still run. But it will give you incorrect results—false negatives. You will erroneously conclude that there is no similarity between your query sequence and the selected database. You should have used blastn, tblastn or tblastx depending on your database and the expected evolutionary distance between your query and the sequences you are comparing against.
SequenceServer automatically chooses the right algorithm depending on your query and database sequence types
So, if you’re running BLAST locally or at NCBI, you need to know the type of query sequence and the type of database sequence. Think carefully before clicking.
However, if you’re using SequenceServer, no need to worry. SequenceServer automatically chooses the appropriate algorithm. Indeed, it has an “automagic” selection mechanism that identifies query type and database type, and selects the BLAST algorithm that will work best. You can focus on the science and avoid costly mistakes.
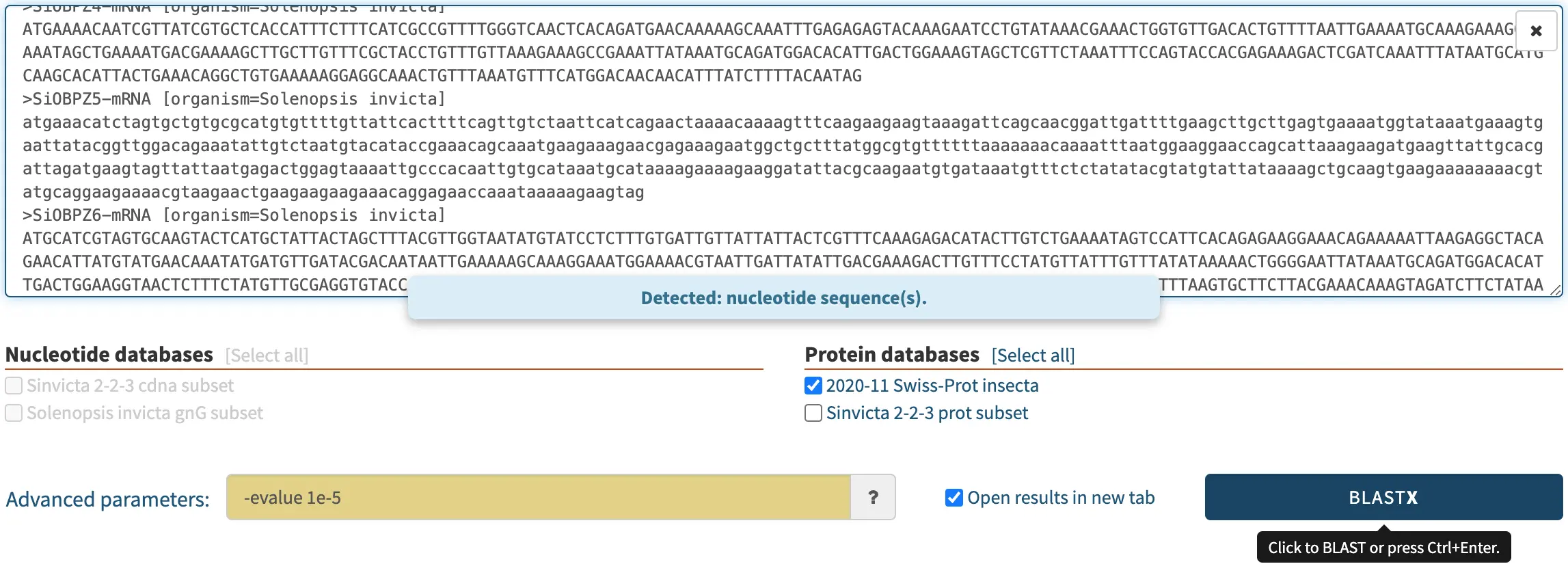
In the screenshot below, a biologist pasted some nucleotide sequences as the query, and selected a protein database. SequenceServer auto-detected this and consequently selected BLASTX, the only algorithm appropriate for comparing nucleotide sequences to a protein database.
blastn vs. tblastx: two options for comparing nucleotide sequences
Things are a bit more complex if you search with nucleotide query sequences against nucleotide databases. You have a choice between blastn and tblastx. Why are there two algorithms that seemingly do the same thing? What are the tradeoffs, and which should you choose?
Algorithmic differences between blastn and tblastx
In short, blastn does comparisons in nucleotide space. It compares nucleotides directly. It does this using the forward sequence, and the reverse-complement sequence.
In contrast, tblastx performs its comparisons in the world of amino-acid sequences. For that, tblastx translates the nucleotide query sequence into amino-acid sequences using all six possible reading frames (three forward and three reverse-complement). And tblastx does the same thing with the nucleotide database, translating it into all six possible translated amino-acid sequences. Thus, each query sequence is effectively compared to the database sequence in thirty-six directions.
Tradeoffs between blastn and tblastx
The algorithmic differences between blastn and tblastx create multiple tradeoffs:
blastnis faster because it makes far fewer comparisons, and each comparison is more straightforward thantblastx.blastnis more precise for highly similar nucleotide sequences.tblastxis more sensitive for divergent sequences. Indeed, it can better detect similarity among distantly related sequences thanblastn. This is because nucleotides degenerate faster than amino acids (because there are 4 * 4 * 4 = 64 possible codons for 20 amino acids plus the “stop signal”, there is some redundancy; thus, different nucleotide sequences can encode identical amino acid sequences).- Only use
tblastxfor protein-coding genes. Remember that translating nucleotide sequences into protein sequences isn’t always reasonable, for example, non-coding RNAs, conserved non-coding elements, or primer sequences.
Conclusion
In conclusion, it’s crucial to choose the right algorithm for your data types and question. SequenceServer will automatically choose what works for the sequence types you’re entering. But if you’re running BLAST locally or at NCBI, you must carefully think through which types of query and database sequences you’re comparing.

For specific applications, additional adjustments are needed. For example,
- for verifying primer sequences, you’ll want to use
blastnand tweak other parameters such as word size and the E-value threshold. - to identify protein-coding genes that are orthologous between species for which you have protein-coding genesets, you’ll want to use
blastp. But if you only have transcriptome assemblies,tblastxmay be more appropriate.